CMI researchers from Idaho State University, Penn State, and Lawrence Livermore National Laboratory conducted the activity for this highlight
Innovation
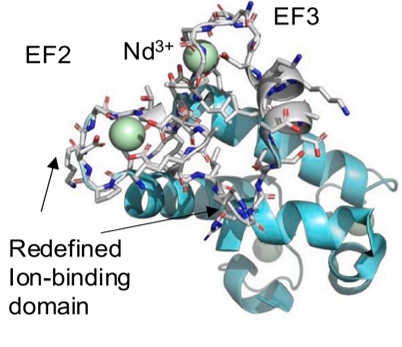
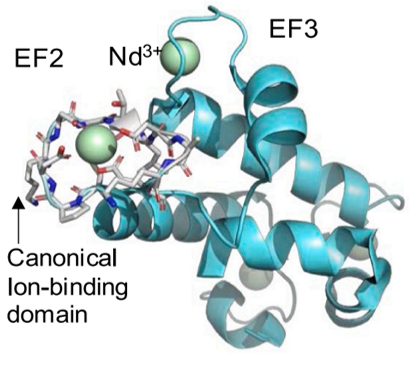
Developed and validated a biomolecular simulation platform for explaining structural underpinnings of selective binding of REEs to the coordination sites (EF hands) of the monomeric bacterial protein lanmodulin.
Achievement
Determined the structural factors underlying the interplay between long- and short-range forces within lanmodulin protein variants that drive differential binding to six REEs. We demonstrated good agreement between computationally predicted binding energies and experimental binding data.
Significance and Impact
- Important first step in achieving a molecular-level understanding of the complex repertoire of intra-molecular forces in lanmodulin and its variants that facilitate differential binding among REEs.
- Next Steps: Extend model to dimeric lanmodulins and use our improved understanding of the biochemical rules to design more selective and shorter lanmodulin variants.
Hub Target Addressed
Developing highly selective separation from complex sources.