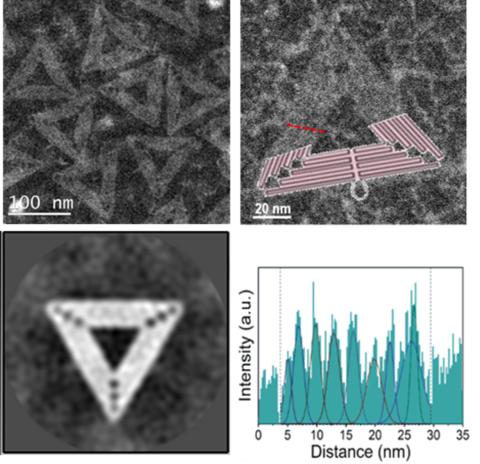
DNA origami refers to the technique of folding of long “scaffold” strands of single-stranded circular DNA molecules held together using short “staple” strands to create various two-or three-dimensional shapes at the nanometer scale. Such nanostructures are potentially useful in a number of applications, including shape-specific foundation to form plasmonic devices. Due to their chemical makeup, the DNA origami nanostructures are difficult to image, with their key structural details typically resolved by using atom force microscopy. Scanning transmission electron microscopy (HAADF-STEM) is used to directly visualize the helices and the seams on the scaffold contacts comprising the DNA origami triangles, without the use of chemistry-altering negative staining. A2D average image was obtained by using a specialized routine employed by the cryo-electron microscopy community, for the first time. An advantage of STEM analysis of DNA origami is the implementation of analytical spectroscopy techniques to provide spatio-chemical characterization of these nanostructures. Now that we have a good understanding of structural detail of DNA origami structures and their surface chemistry, we can utilize this knowledge for precise and controlled metallization of these nanostructures.
A. Londono-Calderon, Md. M. Hossen, P. E. Palo, L. Bendickson, S. Vergara, M. Nilsen-Hamilton, A. C. Hillier, T. Prozorov “Imaging of Unstained DNA Origami Triangles with Electron Microscopy”, Small Methods, 3, 1900393 (2019). DOI: 10.1002/smtd.201900393.
